Basic Immunoglobulin structure
A. Heavy and Light Chains
All immunoglobulins have a four chain structure as their basic unit. They are composed of two identical light chains (23kD) and two identical heavy chains (50-70kD)
B. Disulfide bonds
1. Inter-chain disulfide bonds - The heavy and light chains and the two heavy chains are held together by inter-chain disulfide bonds and by non-covalent interactions The number of inter-chain disulfide bonds varies among different immunoglobulin molecules.
2. Intra-chain disulfide binds - Within each of the polypeptide chains there are also intra-chain disulfide bonds.
C. Variable (V) and Constant (C) Regions
After the amino acid sequences of many different heavy chains and light chains were compared, it became clear that both the heavy and light chain could be divided into two regions based on variability in the amino acid sequences. These are the:
1. Light Chain - VL (110 amino acids) and CL (110 amino acids)
2. Heavy Chain - VH (110 amino acids) and CH (330-440 amino acids)
D. Hinge Region
This is the region at which the arms of the antibody molecule forms a Y. It is called the hinge region because there is some flexibility in the molecule at this point.
E. Domains
Three dimensional images of the immunoglobulin molecule show that it is not straight as depicted in Figure 2A. Rather, it is folded into globular regions each of which contains an intra-chain disulfide bond (figure 2B-D). These regions are called domains.
1. Light Chain Domains - VL and CL
2. Heavy Chain Domains - VH, CH1 - CH3 (or CH4)
F. Oligosaccharides
Carbohydrates are attached to the CH2 domain in most immunoglobulins. However, in some cases carbohydrates may also be attached at other locations.
IV. STRUCTURE OF THE VARIABLE REGION
A. Hypervariable (HVR) or complementarity determining regions (CDR)
Comparisons of the amino acid sequences of the variable regions of immunoglobulins show that most of the variability resides in three regions called the hypervariable regions or the complementarity determining regions as illustrated in Figure 3. Antibodies with different specificities (i.e. different combining sites) have different complementarity determining regions while antibodies of the exact same specificity have identical complementarity determining regions (i.e. CDR is the antibody combining site). Complementarity determining regions are found in both the H and the L chains.
B. Framework regions
The regions between the complementarity determining regions in the variable region are called the framework regions (Figure 3). Based on similarities and differences in the framework regions the immunoglobulin heavy and light chain variable regions can be divided into groups and subgroups. These represent the products of different variable region genes.
 Figure 3 Structure of the variable region framework regions
Figure 3 Structure of the variable region framework regions V. IMMUNOGLOBULIN FRAGMENTS: STRUCTURE/FUNCTION RELATIONSHIPS
Immunoglobulin fragments produced by proteolytic digestion have proven very useful in elucidating structure/function relationships in immunoglobulins.
A. Fab
Digestion with papain breaks the immunoglobulin molecule in the hinge region before the H-H inter-chain disulfide bond Figure 4. This results in the formation of two identical fragments that contain the light chain and the VH and CH1 domains of the heavy chain.
Antigen binding - These fragments were called the Fab fragments because they contained the antigen binding sites of the antibody. Each Fab fragment is monovalent whereas the original molecule was divalent. The combining site of the antibody is created by both VH and VL. An antibody is able to bind a particular antigenic determinant because it has a particular combination of VH and VL. Different combinations of a VH and VL result in antibodies that can bind a different antigenic determinants.
B. Fc
Digestion with papain also produces a fragment that contains the remainder of the two heavy chains each containing a CH2 and CH3 domain. This fragment was called Fc because it was easily crystallized.
 Figure 4 Immunoglobulin fragments: Structure/function relationships
Figure 4 Immunoglobulin fragments: Structure/function relationships Effector functions - The effector functions of immunoglobulins are mediated by this part of the molecule. Different functions are mediated by the different domains in this fragment (Figure 5). Normally the ability of an antibody to carry out an effector function requires the prior binding of an antigen; however, there are exceptions to this rule.
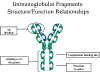 Figure 5 Immunoglobulin fragments: Structure function relationships
Figure 5 Immunoglobulin fragments: Structure function relationships C. F(ab')2
Treatment of immunoglobulins with pepsin results in cleavage of the heavy chain after the H-H inter-chain disulfide bonds resulting in a fragment that contains both antigen binding sites (Figure 6). This fragment was called F(ab')2 because it was divalent. The Fc region of the molecule is digested into small peptides by pepsin. The F(ab')2 binds antigen but it does not mediate the effector functions of antibodies.
See this message from God.